Principle components analysis (PCA) with scikit-learn¶
scikits-learn is a premier machine learning library for python, with a very easy to use API and great documentation.
In [1]: import mdtraj as md
In [2]: from sklearn.decomposition import PCA
Lets load up our trajectory. This is the trajectory that we generated in the “Running a simulation in OpenMM and analyzing the results with mdtraj” example.
In [3]: traj = md.load('ala2.h5')
In [4]: print traj
<mdtraj.Trajectory with 100 frames, 22 atoms>
Create a two component PCA model, and project our data down into this reduced dimensional space. Using just the cartesian coordinates as input to PCA, it’s important to start with some kind of alignment.
In [5]: pca1 = PCA(n_components=2)
In [6]: traj.superpose(traj, 0)
Out[6]: <mdtraj.Trajectory with 100 frames, 22 atoms at 0x10ee6a450>
In [7]: reduced_cartesian = pca1.fit_transform(traj.xyz.reshape(traj.n_frames, traj.n_atoms * 3))
In [8]: print reduced_cartesian.shape
(100, 2)
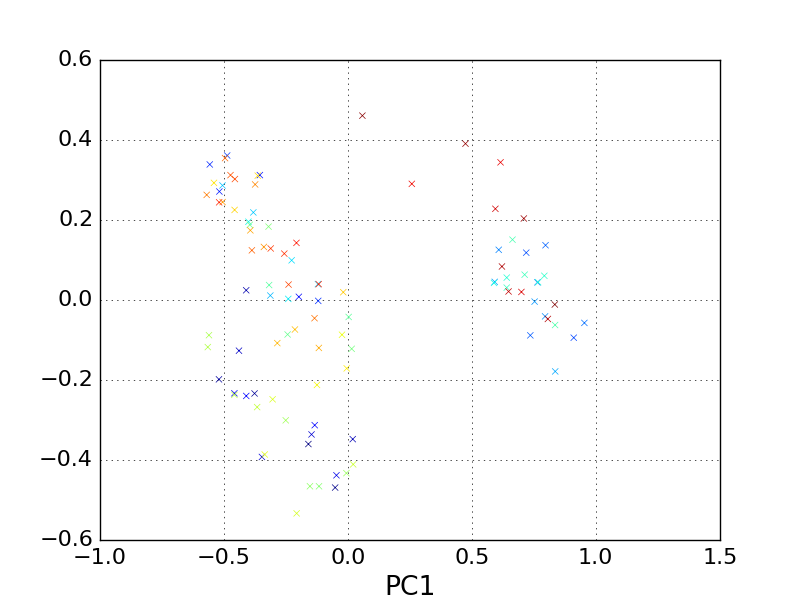
Now we can plot the data on this projection.
In [9]: import matplotlib.pyplot as pp
In [10]: pp.figure()
Out[10]: <matplotlib.figure.Figure at 0x10e66e290>
In [11]: pp.scatter(reduced_cartesian[:, 0], reduced_cartesian[:,1], marker='x', c=traj.time)
Out[11]: <matplotlib.collections.PathCollection at 0x10e5cf190>
In [12]: pp.xlabel('PC1')
Out[12]: <matplotlib.text.Text at 0x10e6d0610>
In [13]: pp.ylabel('PC2')
Out[13]: <matplotlib.text.Text at 0x10e6d7650>
In [14]: pp.title('Cartesian coordinate PCA: alanine dipeptide')
Out[14]: <matplotlib.text.Text at 0x10e6c3590>
In [15]: cbar = pp.colorbar()
In [16]: cbar.set_label('Time [ps]')
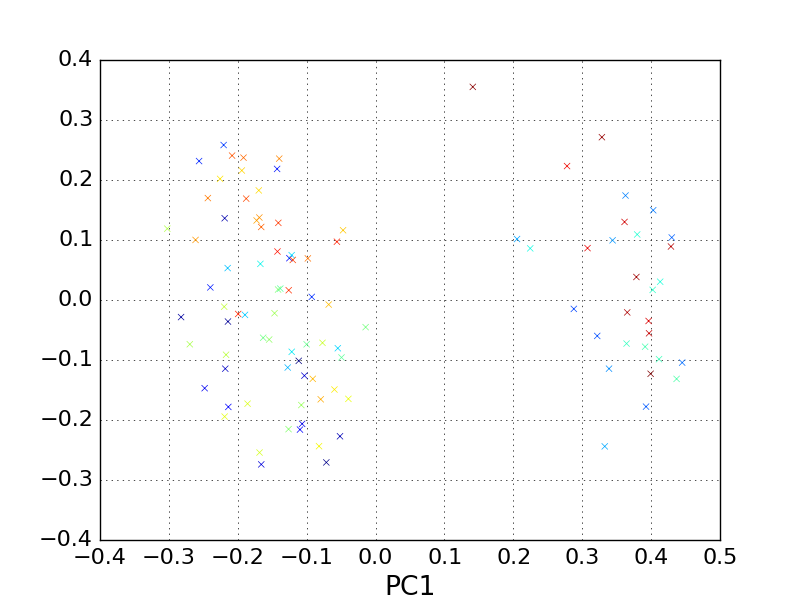
Lets try cross-checking our result by using a different feature space that isn’t sensitive to alignment, and instead to “featurize” our trajectory by computing the pairwise distance between every atom in each frame, and using that as our high dimensional input space for PCA.
In [17]: pca2 = PCA(n_components=2)
In [18]: from itertools import combinations
this python function gives you all unique pairs of elements from a list
In [19]: atom_pairs = list(combinations(range(traj.n_atoms), 2))
In [20]: pairwise_distances = md.geometry.compute_distances(traj, atom_pairs)
In [21]: print pairwise_distances.shape
(100, 231)
In [22]: reduced_distances = pca2.fit_transform(pairwise_distances)
In [23]: import matplotlib.pyplot as pp
In [24]: pp.figure()
Out[24]: <matplotlib.figure.Figure at 0x10e6fac90>
In [25]: pp.scatter(reduced_distances[:, 0], reduced_distances[:,1], marker='x', c=traj.time)
Out[25]: <matplotlib.collections.PathCollection at 0x11080e250>
In [26]: pp.xlabel('PC1')
Out[26]: <matplotlib.text.Text at 0x10e6e9350>
In [27]: pp.ylabel('PC2')
Out[27]: <matplotlib.text.Text at 0x10f07e190>
In [28]: pp.title('Pairwise distance PCA: alanine dipeptide')
Out[28]: <matplotlib.text.Text at 0x10eeb4390>
In [29]: cbar = pp.colorbar()
In [30]: cbar.set_label('Time [ps]')